
Synthetic formation of a type of subcellular membraneless compartment in E. coli
upon overexpression of disordered structural proteins such as spider silk and resilin.
Shaopeng Wei
Graduate Student, Shanghai Jiao Tong University
Over the past decades, scientists are fascinated by the subcellular organelles with sophisticated structures and biological functions. Taking inspiration from nature, scientists have been seeking to construct artificial organelles for synthetic applications. Indeed, we can now reprogram naturally occurring organelles with on-demand functions in living cells. However, it is still a challenge to create an organelle from scratch.
Now, the explosion of studies involving membraneless organelles offers us an alternative way to construct subcellular compartments. Membraneless organelles, a newly defined type of cellular compartment, are unlike their membrane-bound counterparts, and form via liquid–liquid phase separation (LLPS) of biomacromolecules such as proteins and nucleic acids within eukaryotic cells. However, there is no direct evidence for the existence of such organelles in prokaryotes. The LLPS compartmentalization mechanism has motivated us to develop a strategy for designing synthetic, genetically encoded mimics, and indeed, some artificial membraneless organelles have been made with the natural or modified LLPS-prone biomacromolecules in eukaryotes. Yet this strategy has not been successful in prokaryotes as one major obstacle is that eukaryotic LLPS proteins are highly aggregation-prone and difficult to express.
Our lab has long been focusing on the design and biosynthesis of protein polymers and biomaterials1, and my first project was actually the production of recombinant spider silk proteins in the bacterial cell factory Escherichia coli, following the endeavors of my advisors and lab seniors2,3. At first, I came up with an idea to label the spider silk protein with a green fluorescent protein, aiming to “see” the silk production levels in the bacterial cells by measuring fluorescence. Unfortunately, this did not always work reliably. The lucky thing was that my supervisor advised me to investigate the intracellular distribution of the spider silk proteins via fluorescence microscopy. Interestingly, the recombinant spider silk proteins spatially localized near the pole regions of E. coli cells (see Figure 1 below), which shocked me, simply because the silk proteins were expressed in a soluble form instead of insoluble inclusion bodies that are commonly seen in prokaryotic expression systems. Then, why and how the silk proteins condense in the living cells? It was later made clear that our spider silk proteins, natively unfolded and disordered with low-complexity sequences, share common features with the eukaryotic LLPS proteins that are naturally involved in membraneless organelle formation. Then we verified that LLPS underlay the formation of silk condensates within E. coli cells, and demonstrated functionalization of the synthetic membraneless compartments with biocatalytic activities4.Furthermore, we found that other disordered structural proteins (e.g. resilin) formed similar compartments inside the E. coli cells, indicating applicability of our compartmentalization strategy with heterologous structural proteins. In addition, the bacterial cell physiology was not substantially altered with the emergence of subcellular liquid condensates, indicating their good potential for biotechnological applications.
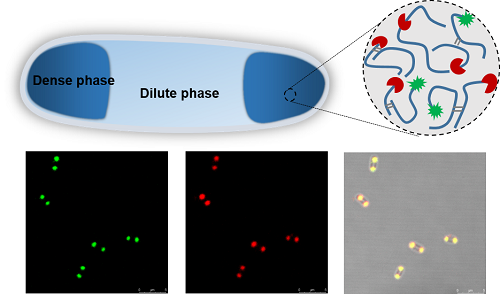
Figure 1: Formation and functionalization of subcellular compartments upon silk expression in E. coli.
In summary, our work provides a new compartmentalization strategy in the model prokaryotic organism E. coli, which not only demonstrates the feasibility of constructing artificial membraneless organelles within cells of prokaryotes, but also opens the door to broad applications in metabolic engineering and synthetic biology in the near future.
References
1. Qian, Z. G., Pan, F., Xia, X. X. Synthetic biology for protein-based materials. Curr.Opin. Biotechnol. 65, 197-204 (2020).
2. Xia, X. X. et al. Native-sized recombinant spider silk protein produced in metabolically engineered Escherichia coli results in a strong fiber. Proc. Natl. Acad. Sci. USA 107, 14059-14063 (2010).
3. Yang, Y. X., Qian, Z. G., Zhong, J. J., Xia, X. X. Hyper-production of large proteins of spider dragline silk MaSp2 by Escherichia coli via synthetic biology approach.Process Biochem. 51, 484-490 (2016).
4. Wei, S. P. et al. Formation and functionalization of membraneless compartments in Escherichia coli. Nat. Chem. Biol.(2020).
https://www.nature.com/articles/s41589-020-0579-9.

Address: 800 Dongchuan RD. Minhang District, Shanghai, China
School of Life Sciences and Biotechnology, SJTU Copyright © 2017 沪交ICP备05029. All Rights Reserved.


